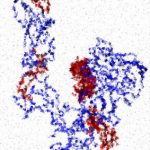
Hepcidin and its selective behavior toward SARS-CoV2 spike states
We analyzed the interactions of free and bound forms of spike with hepcidin (HPC), the major hormone in iron regulation,…
Covid-19 hits ethnic groups
Covid-19 hits Iranian ethnic groups differentially In current work we discuss the effect of worldwide polymorphism of ACE2 protein on…
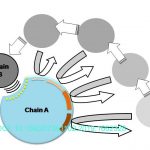
Protein Target Finding
Our observations indicate that the mechanical properties of protein binding sites have a decisive impact on determining the dimer stability….…